Animal Molecular Genetics Lab

Principle Investigator: Catherine Ernst
Anthony Hall Rooms 2209 and 3330 | 474 S. Shaw Lane | East Lansing, MI 48824




Anthony Hall Rooms 2209 and 3330 | 474 S. Shaw Lane | East Lansing, MI 48824


The Animal Molecular Genetics Laboratory focuses on identification and evaluation of genes and genetic markers associated with genetic improvement of production traits in pigs and other meat animal species. Much of our research is at the interface of structural and functional genomics, including a genetical genomics project to integrate genetic marker and gene expression data to identify genes controlling skeletal muscle and fat deposition, and their relationship to growth, carcass composition and meat quality in pigs. This effort applies both RNAseq and miRNAseq approaches in order to reveal gene regulatory networks and pathways. The lab also uses functional genomics approaches including transcriptome sequencing and assessment of DNA methylation patterns to study pig skeletal muscle development, and stress response in pigs. Additional projects include discovery and evaluation of RNA editing sites, and cataloging DNA methylation patterns for various pig tissues.
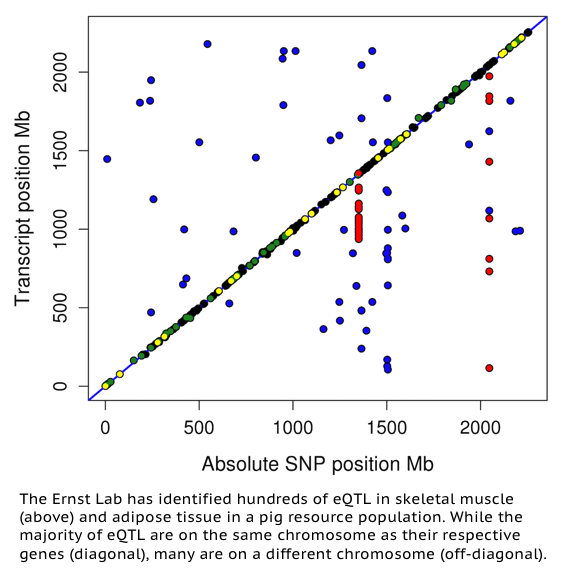
The application of genomic technologies is enhancing pig genetic improvement. Genome-wide analyses have revealed genomic regions harboring single nucleotide polymorphisms (SNPs) contributing significant portions of phenotypic trait variation. However, the molecular mechanisms underlying these contributions remain unclear. We aim to identify expression quantitative trait loci (eQTL) in the F2 generation of the Michigan State University Pig Resource Population (MSUPRP) to elucidate the genetic architecture of gene expression variation and gain insight on the genetic contribution underlying differences in polygenic traits. Associating gene expression profiles with genome-wide SNP markers reveals both local and distal regulators of gene expression. Local eQTL identify candidate loci directly influencing the expression of the associated gene and thus infers direct cause of variation in gene expression. Distal eQTL identify candidate gene-gene interactions where the expression of one gene may affect the expression of distant genes through transcriptional co-regulation. As sequencing costs go down, eQTL studies have the potential to be incorporated into expression-assisted evaluations in animal breeding by the use of genetical genomic selection of both phenotypic QTL and expression QTL. Our lab is currently conducting eQTL studies utilizing genotype and transcriptomic data (mRNA and miRNA) from adipose and skeletal muscle tissues to identify genomic loci influencing gene expression and economically-important pig-production phenotypes related to growth, meat quality, and carcass composition.
Recent Publications & Presentations:
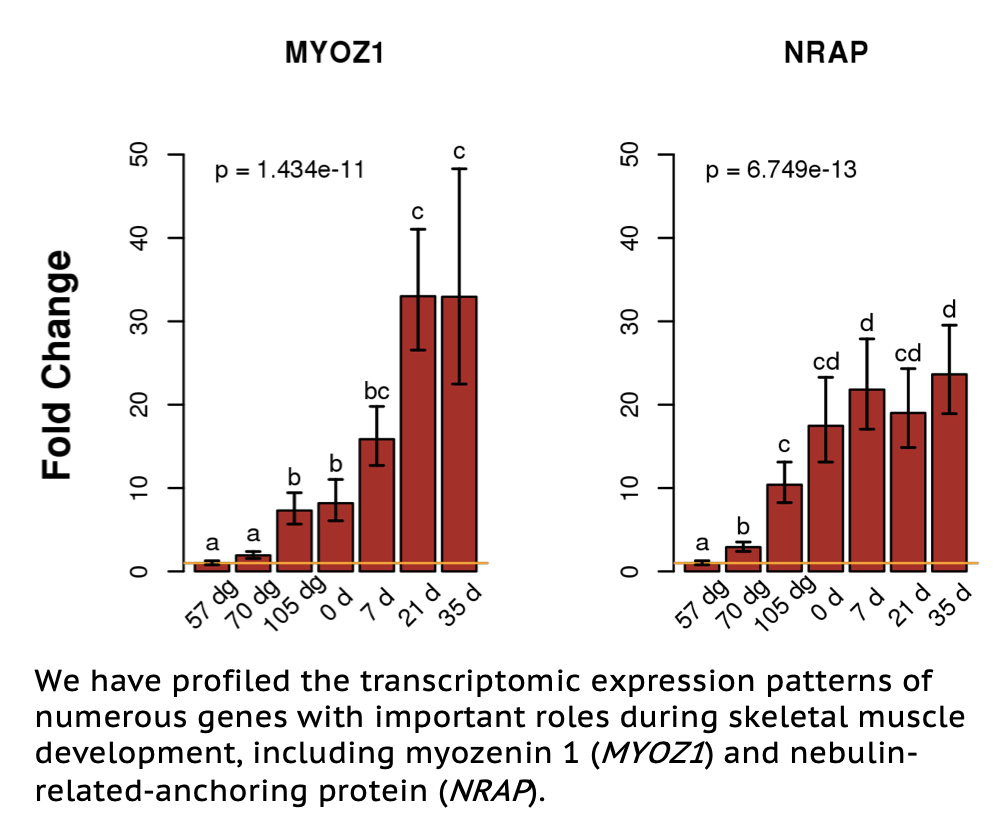
The Ernst lab has had a long-term research interest in the mechanisms that control skeletal muscle growth and development. Development, growth and function of skeletal muscle are dynamic processes. Skeletal muscle ultimately becomes meat, the product of pig production, and skeletal muscle accounts for 40-65% of carcass weight. The number of muscle fibers in pigs, as well as most mammals, is determined prenatally. Development of skeletal muscle fibers during fetal development occurs in two waves during which myoblasts proliferate and fuse to form new fibers. Primary fibers form de novo and secondary fibers form around primary fibers. In pigs these processes take place at approximately 30-60 and 54-90 days of gestation, respectively. Our lab has collected skeletal muscle samples from an ontogeny of fetal and post-natal ages. RNA from these samples has been evaluated for transcript abundance and differential expression between ages using expression microarrays and real time quantitative PCR (qPCR). Ongoing work on this project involves examining transcript abundance using RNA-seq and microRNA-seq, as well as applying whole-genome bisulfite sequencing and other epigenetic approaches to further annotate the skeletal muscle transcriptome.
Recent Publications & Presentations:
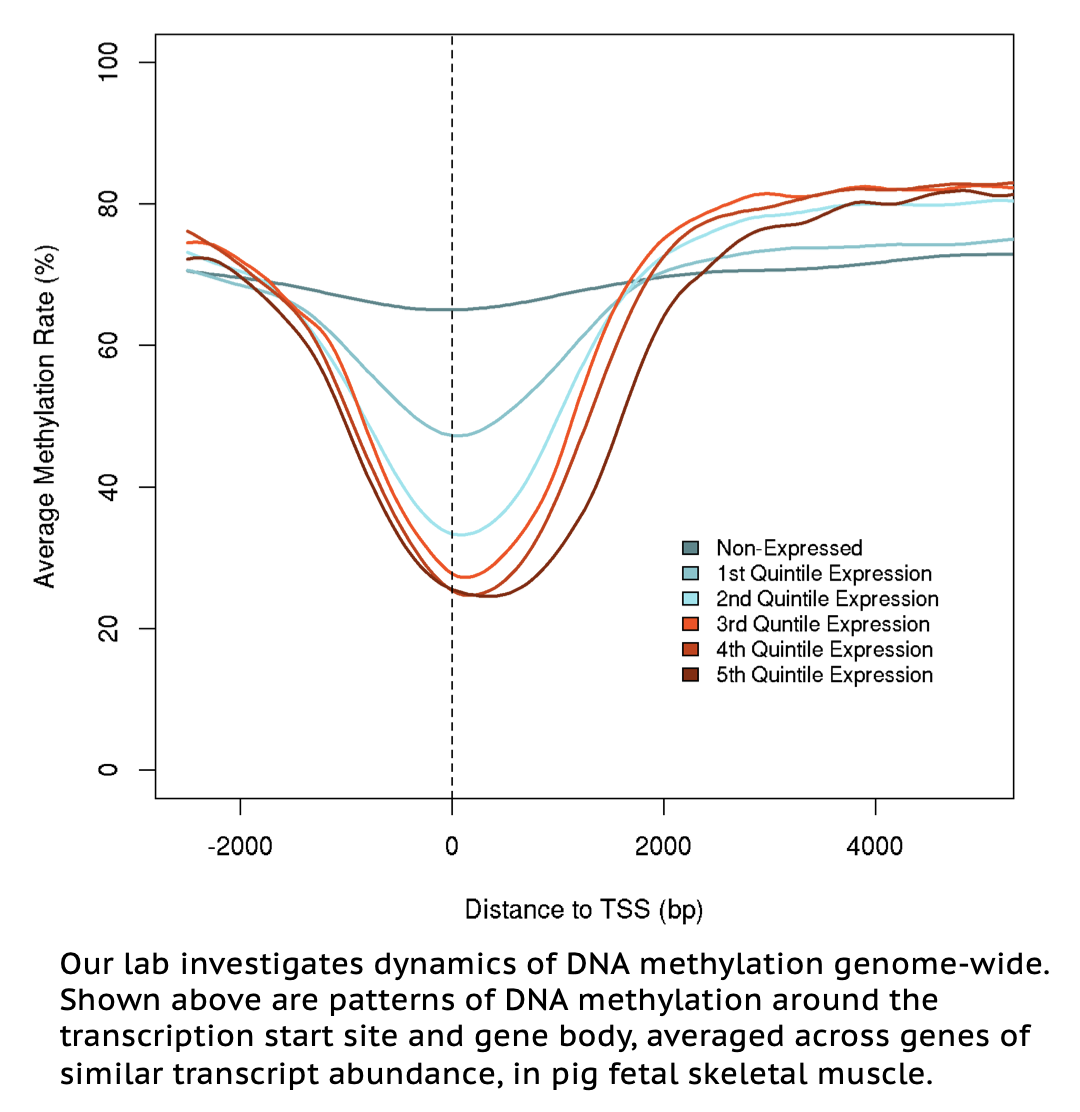
Epigenetic processes result in heritable changes in gene expression without changes in DNA sequence. DNA methylation—the most studied epigenetic modification to DNA—involves the enzymatic addition of a methyl group to cytosine bases in DNA, and in mammals occurs almost exclusively at CpG dinucleotides. CpG methylation at proximal and distal gene regulatory regions—including promoters, enhancers, and insulators—is strongly associated with transcription, and as such has been shown to play important roles in tissue-specific gene expression, early development, and genomic imprinting. DNA methylation patterns in mammals are also heavily influenced by environmental perturbations; aberrant DNA methylation in response to stressors has been associated with adverse life outcomes. The Ernst lab is currently using Whole-Genome Bisulfite Sequencing (WGBS) to assess DNA methylation patterns across a diverse set of adult and fetal pig tissues as a means to identify novel sites of transcriptional regulation. The lab also utilizes WGBS to assess methylation dynamics in pig immune cells in response to stressors.
Recent Publications & Presentations:
Among the complex ways in which animals can regulate the expression of transcripts, endogenous RNA editing has been shown to be widespread, catalyzed primarily by the nuclear expressed family of adenosine deaminase acting on RNA (ADAR) proteins that perform single adenosine to inosine transitions along premature RNA transcripts. It remains unknown to what degree transcriptome-wide RNA editing exists in livestock species and its overall effect on controlling complex traits. We have built bioinformatic software to streamline the RNA editing detection process from whole genome sequencing and RNA-seq data and have begun to apply it to the pig model, observing inherent associations between candidate RNA editing sites and swine-specific SINE retrotransposons. We are evaluating the stability and repeatability of RNA edited sites among pigs, as well as assessing the spatial and temporal variation of edited sites. This work is crucial to understanding the extent to which RNA editing contributes to single nucleotide variation among swine transcriptomes, and how this unique mechanism of transcriptional regulation contributes to complex phenotypes.
Recent Publications & Presentations:
Our lab uses several different pig populations to evaluate genetic variation associated with candidate genes. Most of the genes we study are selected based on their map position within regions of quantitative trait loci (QTL) identified for phenotypic traits in our Duroc x Pietrain resource population. Single nucleotide polymorphisms (SNPs) in candidate genes are either identified in our lab or obtained from the literature, and SNP genotypes are associated with phenotypes. Recent genes that we have evaluated include IGFBP2, CRHR2, PRKAG3 and LEPR. For some genes, we also evaluate transcript abundance in longissimus dorsi muscle, subcutaneous fat and/or liver samples from our resource population pigs.
Recent Publications & Presentations:
I started my research program at MSU in 1997 and I am currently a professor of Animal Science. I also hold partial administrative appointments as Associate Chair for Graduate Training and Research in the Department of Animal Science, and as Director of the MSU interdepartmental Genetics and Genome Sciences Graduate Program. Nationally, I serve as Pig Genome Co-Coordinator for the USDA National Animal Genome Research Program (NRSP-8). Before starting my faculty position, I received my MS from Iowa State University and my PhD from The Ohio State University, and did postdoctoral research at the USDA Meat Animal Research Center in Clay Center, NE. In addition to research, I co-teach an undergraduate genetics course, and I facilitate a graduate scientific writing course and a graduate seminar course.
I have been the lab manager for the Ernst lab since 1997 and am also lab manager for two other faculty members. I received my BS and MS from the University of Kentucky. In addition to conducting research, I am responsible for overseeing the routine functioning of the laboratory as well as training students. When I’m not at work, I’m busy trying to keep up with my twin girls.
I’m currently a PhD candidate in the Department of Animal Science’s Animal Breeding and Genetics Group, and a recipient of the USDA NIFA National Needs Fellowship (2013 – 2016). I earned a B.S. in Animal Science from MSU in 2013, and my previous research experience studied dairy cattle feeding behavior at the William H. Miner Agricultural Research Institute in Chazy, NY (Summer 2013). My current research focuses on understanding the effects of microRNA regulation on meat quality and carcass phenotypes in pigs utilizing GWAS, regulatory network analysis, and molecular genetics techniques. When I’m not working on research, I enjoy spending time with my husband, friends, and family, and adventuring new parks and trails with my dog, Willow.
The majority of my research involves analyzing whole-genome DNA methylation patterns in pig adult and fetal tissues to enhance discovery of regulatory elements and increase functional annotation of the porcine genome. My other projects involve studying RNA editing dynamics during pig development, as well as how the epigenome of livestock species is impacted by stressors experienced in production settings. I have previously been funded under a USDA NIFA National Needs Training Grant (2016-Spring 2019), and I am currently funded by a USDA NIFA Predoctoral Fellowship. Additionally, I am actively involved in genetics-related community outreach and engagement in the Greater Lansing area. In my free time, I am usually triathlon training or sitting on my couch with a book or crossword puzzle.
I am currently an undergraduate Animal Science major. My research involves profiling the expression of genes of interest in cultured mouse skeletal muscle cells to define their effects on myogenesis. In my free time, I enjoy running, attending Spartan sporting events, running sled dogs, and spending time with my friends.




















Velez-Irizarry D., Casiro S., Daza K.R., Bates R.O., Raney N.E., Steibel J.P. and Ernst C.W. 2019. Genetic control of longissimus dorsi muscle gene expression variation and joint analysis with phenotypic quantitative trait loci in pigs. BMC Genomics. 20(1):3. doi: 10.1186/s12864-018-5386-2
Wurtz, K.E. J.M. Siegford, C.W. Ernst, N.E. Raney, R.O. Bates and J.P. Steibel. 2018. Genome-wide association analyses of lesion counts in group-housed pigs. Anim. Genet. 49(6):628-31. doi: 10.1111/age.12713
Zhao, P., Y. Wu, W. Feng, H. Du, J. Yu, H. Kang, X. Zheng, Z. Wang, G.E. Liu, C.W. Ernst, X. Ran, J. Wang and J.-F. Liu. 2018. Evidence of evolutionary history and selective sweep in the genome of Meishan pig reveals its genetic and phenotypic characterization. Gigascience. 7:1-12. doi: 10.1093/gigascience/giy058
Casiró S, D. Velez-Irizarry, C.W. Ernst, N.E. Raney, R.O. Bates, M.G. Charles and J.P. Steibel. 2017. Genome-wide association study in an F2 Duroc x Pietrain resource population for economically important meat quality and carcass traits. J. Anim. Sci. 95:545-558. doi: 10.2527/jas.2016.1003
Choi, I., R.O. Bates, N.E. Raney and C.W. Ernst. 2017. Association of a corticotropin-releasing hormone receptor 2 (CRHR2) polymorphism with carcass merit, meat quality and stress response traits in pigs. Canadian J. Anim. Sci. 97:536-540. doi: 10.1139/cjas-2016-0168
Daza, K.R., J.P. Steibel, D. Velez-Irizarry, N.E. Raney, R.O. Bates and C.W. Ernst. 2017. Profiling and characterization of a longissimus dorsi muscle microRNA dataset from an F2 Duroc x Pietrain pig resource population. Genom. Data. 13:50-53. doi: 10.1016/j.gdata.2017.07.006
Funkhouser, S.A., R.O. Bates, C.W. Ernst, D. Newcom and J.P. Steibel. 2017. Estimation of genome-wide and locus-specific breed composition in pigs. Translational Anim. Sci. 1:36-44. doi: 10.2527/tas2016.0003
Funkhouser, S.A., J.P. Steibel, R.O. Bates, N.E. Raney and C.W. Ernst. 2017. Evidence for transcriptome-wide RNA editing among Sus scrofa PRE-1 SINE elements. BMC Genomics.18:360. doi: 10.1186/s12864-017-3766-7
Wurtz, K.E. J.M. Siegford, R.O. Bates, C.W. Ernst and J.P. Steibel. 2017. Estimation of genetic parameters for lesion scores and growth traits in group-housed pigs. J. Anim. Sci. 95:4310- 4317. doi: 10.2527/jas2017.1757.
Bernal-Rubio, Y.L., J.L. Gualdron-Duarte, R.O. Bates, C.W. Ernst, D. Nonneman, G.A. Rohrer, D.A. King, S.D. Shackelford, T.L. Wheeler, R.J.C. Cantet and J.P. Steibel. 2016. Meta-analysis of genome-wide association from genetic prediction models. Anim. Genet. 47:36-48.
Gualdron Duarte, J.L., R.J. Cantet, Y.L. Bernal-Rubio, R.O. Bates, C.W. Ernst, N.E. Raney, A. Rogberg-Muñoz and J.P. Seibel. 2016. Refining genomewide association for growth and fat deposition traits in an F2 pig population. J. Anim. Sci. 94:1387-1397.
Additional publications may be found at ResearchGate, PubMed, and Google Scholar